Establishing gap gene expression
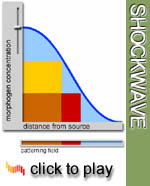 Bicoid
regulates multiple gap genes in a concentration dependent manner. In
this way, the Bicoid concentration gradient is transformed into a spatial
pattern. A gene X may be active only at high concentrations of Bicoid,
whereas gene Y, which has a stronger binding site, is active also at
lower concentrations, and therefore at greater distance from the source.
Manipulating the concentration of Bicoid in the embryo by genetic means
leads to a complete shift of embryonic cell fates anteriorly (in case
of decreased bicoid concentration) or posteriorly (in case of an increase).
Today we know that these fate shifts are not exactly proportional to
the variation in local concentration. Other factors play additional
roles in target gene regulation.
Bicoid
regulates multiple gap genes in a concentration dependent manner. In
this way, the Bicoid concentration gradient is transformed into a spatial
pattern. A gene X may be active only at high concentrations of Bicoid,
whereas gene Y, which has a stronger binding site, is active also at
lower concentrations, and therefore at greater distance from the source.
Manipulating the concentration of Bicoid in the embryo by genetic means
leads to a complete shift of embryonic cell fates anteriorly (in case
of decreased bicoid concentration) or posteriorly (in case of an increase).
Today we know that these fate shifts are not exactly proportional to
the variation in local concentration. Other factors play additional
roles in target gene regulation. Gap
genes are regulated by maternal genes, which also feed upon each other.
Gap
genes are regulated by maternal genes, which also feed upon each other.
Bicoid is a transcription factor; it binds to DNA and regulates downstream genes in the anterior region of the embryo, like the zygotic hunchback and the anterior giant domain. Further anterior genes under the control of bicoid are orthodenticle, buttonhead and empty spiracles (not shown here). In addition, Bicoid can also act as a translational repressor of maternal caudal RNA. Whereas caudal RNA is ubiquitously distributed in the early embryo, Caudal protein is restricted to a posterior gradient inverse to the bicoid protein distribution.
This double function of Bicoid is carried out by two different proteins for the posterior system: Nanos is a translational regulator and represses the translation of maternal hunchback mRNA in conjunction with the gene product of pumilio, thereby restricting Hunchback protein distribution to the anterior region of the embryo. Caudal protein acts as a transcription factor to regulate gap gene expression.
Hunchback is a transcription factor and assists bicoid in the regulation of anterior zygotic genes. The expression of Krüppel for example requires both the presence of hunchback and bicoid.
In the absence of nanos, Hunchback protein becomes uniformly distributed throughout the egg. The ectopic presence of Hunchback protein in the posterior part of the embryo represses the expression of knirps and the posterior giant domain. This shows that hunchback can act as a repressor of posterior gap genes. This role of hunchback in keeping the anterior half of the embryo void of posterior gene expression can also be observed in embryos lacking the maternal and zygotic hunchback contribution (not shown here).
The posterior gap genes are activated by both bicoid and caudal. The effect of bicoid on the expression of posterior genes is very weak, whereas the absence of caudal leads to a considerable reduction in the expression of knirps and the posterior giant domain, for example. In the absence of both bicoid and caudal, posterior gap genes are completely abolished.
The terminal gap genes tailless and huckebein (not shown here) are activated by the terminal maternal system.
Media list
Genes discussed
|
Gene
|
Gene product
- Domains
|
Function
|
Links
|
|
bicoid
(bcd)
|
transcription factor - homeodomain
|
maternally provided morphogen involved in anterior
patterning of the Drosophila embryo
|
|
|
buttonhead
(btd)
|
transcription
factor - zinc finger
|
transcriptonal
activator that regulates the segmentation of the head
|
|
|
caudal
(cad)
|
transcription
factor - homeodomain
|
plays a role in establishing the posterior domains
of the embryo
|
|
|
empty
spiracles (ems)
|
transcription
factor - homeodomain
|
required for head + brain development
|
|
|
giant (gt)
|
transcription factor - basic leucine zipper
|
gap gene
|
|
|
hunchback
(hb)
|
transcription factor - zinc finger |
gap gene, later required for proper temporal generation
of NB sublineages
|
|
|
knirps (kni)
|
transcription factor - steroid receptor - zinc
finger
|
gap gene, later organizes the development of the second
wing vein
|
|
|
Krüppel (Kr)
|
transcription factor - zinc finger transcriptional
repressor
|
gap gene, later required for proper temporal generation
of NB sublineages
|
|
|
nanos
(nos)
|
translational
repressor - zinc finger
|
targets
Hunchback- and Bicoid-mRNAs to achieve posterior identity
|
|
|
orthodenticle
(otd)
|
transcription
factor - homeodomain - paired-like
|
acts in a combinatorial fashion with the cephalic gap
genes empty spiracles and buttonhead to
assign segmental identities in the head and brain
|
|
| pumilio (pum) | novel posterior group gene | binds and regulates Hunchback mRNA | Interactive Fly |